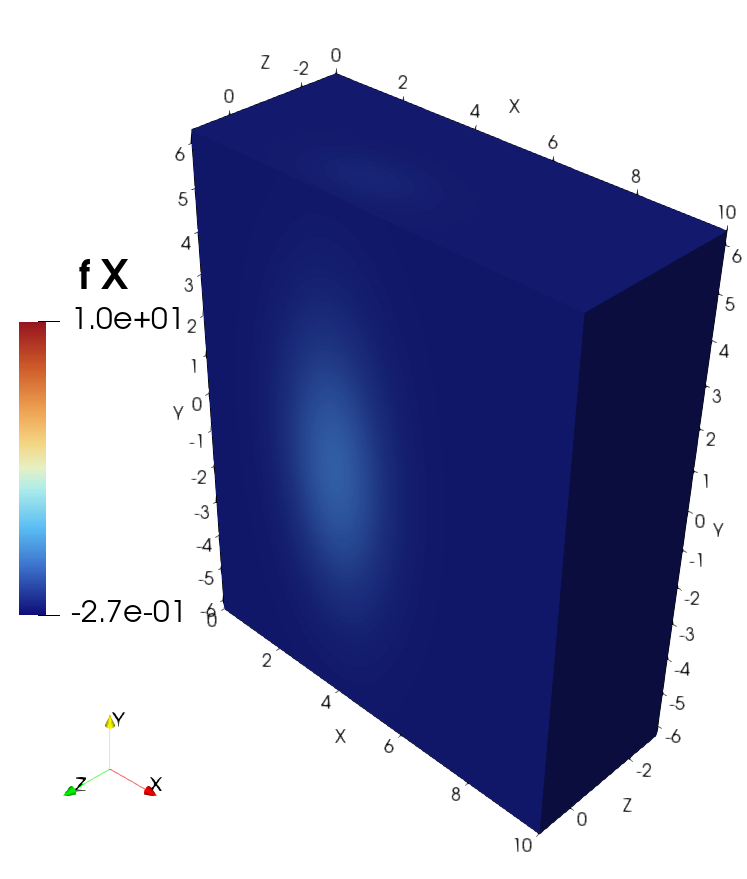
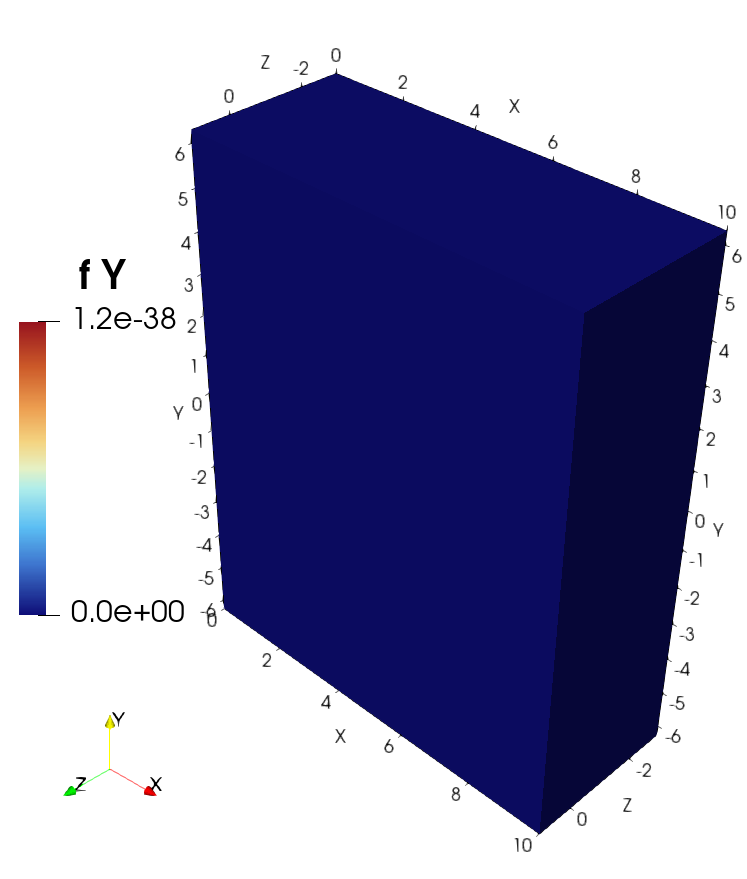
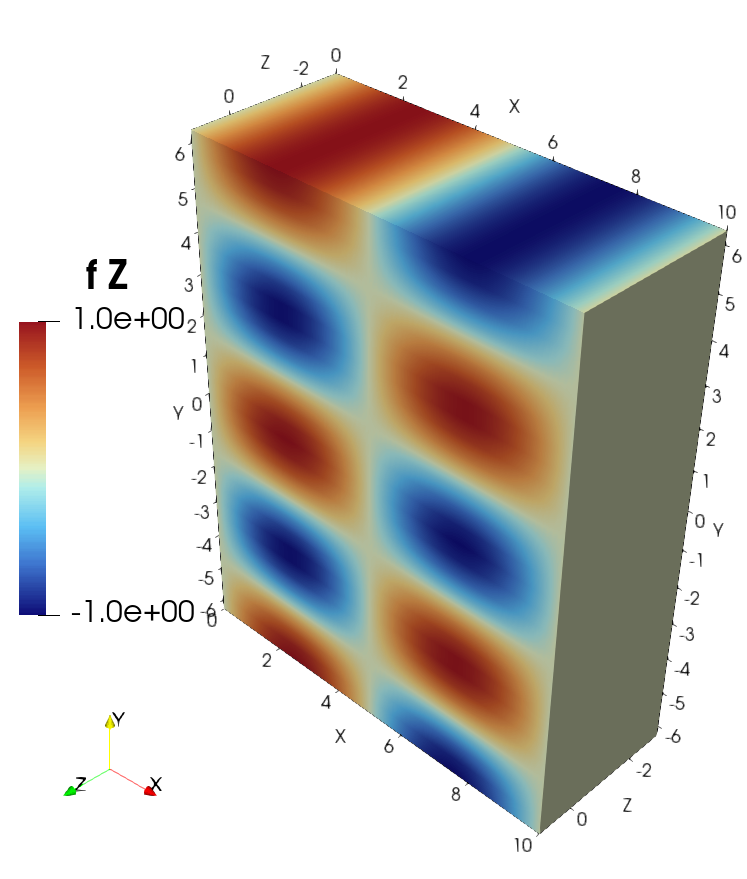
3D example with VTK and ParaView#
In this notebook we show how to solve a 3D problem in parallel, save the solution in HDF5 format and export it to VTK.
We solve the regularized curl-curl problem in 3D with a callable function for the source. In practice, the source could be the interpolation of some experimental data. Besides, we impose periodic boundary conditions everywhere in the boundary.
Let \(\Omega=[0, 10] \times [-2\pi, 2\pi]\times [-3, 1]\). We want to find \(u\in H(\mathrm{curl}, \Omega)\) such that
where \(\mu = \pi\), and \(f\) is a callable function (of the domain coordinates) for which we may not have a symbolic expression.
Since \(\mu>0\) the resulting operator is positive definite and we can use conjugate gradient to solve the system. In the case of the time-harmonic Maxwell problem we have that \(\mu<0\), which yields an indefinite operator and conjugate gradient is no longer guaranteed to converge.
Only for illustration here we use the callable function
and project it from \(H(\mathrm{curl}, \Omega)\) to the corresponding discretized space. This process is analogous with any callable vectorial function.
Step 1 : define the domain and spaces and discretize them#
This is similar to what is done in other examples
[1]:
import numpy as np
from mpi4py import MPI
from sympde.topology import Cube
from sympde.topology import VectorFunctionSpace
from psydac.api.discretization import discretize
comm = MPI.COMM_WORLD # MPI communicator
Omega = Cube('Omega', bounds1=(0, 10), bounds2=(-2*np.pi, 2*np.pi), bounds3=(-3, 1)) # 3D cubic domain
W = VectorFunctionSpace('W', Omega, kind='hcurl') # Hcurl space
ncells = [25, 30, 11] # number of cells in each direction
degree = [3, 2, 2] # B-spline degree in each direction
periodic = [True, True, True] # set periodic BC in every direction
Omega_h = discretize(Omega, ncells=ncells, periodic=periodic, comm=comm)
Wh = discretize(W, Omega_h, degree=degree)
Step 2 : compute the matrix#
We assemble the mass and curl curl matrices.
[2]:
from sympde.topology import elements_of, element_of
from sympde.expr.expr import BilinearForm, LinearForm, integral
from sympde.calculus import dot, curl
from psydac.api.settings import PSYDAC_BACKENDS
from psydac.fem.basic import FemField
backend_language = 'pyccel-gcc'
backend = PSYDAC_BACKENDS[backend_language]
u, v = elements_of(W, names='u, v') #trial and test functions
m = BilinearForm((u, v), integral(Omega, dot(u, v)))
mh = discretize(m, Omega_h, [Wh, Wh], backend=backend)
M = mh.assemble() # Mass matrix
k = BilinearForm((u, v), integral(Omega, dot(curl(u), curl(v))))
kh = discretize(k, Omega_h, [Wh, Wh], backend=backend)
K = kh.assemble() # Curl curl matrix
mu = np.pi
A = K + mu*M # system matrix, positive-definite
Step 3 : compute the right-hand size#
We project a callable function to the vector space and use it to assemble the right-hand side of the system.
[3]:
from psydac.feec.global_geometric_projectors import GlobalGeometricProjectorHcurl
f_callable = [lambda x, y, z: 10*np.exp(-0.5*(x-4)**2 - 1.5*z**2 - 1e-1*y**2),
lambda x, y, z: 0,
lambda x, y, z: np.cos(y)*np.sin(x*np.pi/5)]
proj_Wh = GlobalGeometricProjectorHcurl(Wh, nquads=[p+1 for p in degree]) # get Hcurl projector, nquads is the number of points for the Gauss quadrature.
fh = proj_Wh(f_callable) # project callable to field in Wh
f = element_of(W, name='f') # free variable for the FemField
l = LinearForm(v, integral(Omega, dot(f,v)))
lh = discretize(l, Omega_h, Wh, backend=backend)
rhs = lh.assemble(f=fh) # assmble RHS, f is a free parameter
Step 4 : solve the system#
Solve the system using Conjugate Gradient. Notice that since \(\mu>0\), the matrix is positive definite.
[4]:
from psydac.linalg.solvers import inverse
inv_A = inverse(A, solver='cg', tol=1e-2, maxiter=100) # invert using Conjugate Gradient
u = inv_A @ rhs # array of coefficients
uh = FemField(Wh, coeffs=u) # finite element field, callable
Step 5 : Save results to HDF5 format#
We save the coefficients of the solution and the field used in the right-hand size. This step generates a YAML file containing the space information, and a HDF5 file that stores the coefficients. The fields are saved in parallel. This step does not include an evaluation of the fields on a grid.
[5]:
# Save the results using OutputManager
from psydac.api.postprocessing import OutputManager
import os
os.makedirs('results_regularized_curlcurl', exist_ok=True)
Om = OutputManager(
f'results_regularized_curlcurl/space_info_{Omega.name}.yml',
f'results_regularized_curlcurl/field_info_{Omega.name}.h5',
comm=comm,
save_mpi_rank=True,
mode = 'w'
)
Om.add_spaces(V=Wh)
Om.export_space_info()
Om.set_static() # the fields do not depend on time
save_fields = {
'u': uh,
'f': fh
}
Om.export_fields(**save_fields) # saves the coefficients of the fields
Om.close()
# This generates a YAML file containing the space information, and a HDF5 file that stores the solution coefficients.
# At this point the solution has not been evaluated on any grid.
Step 6 : Evaluate fields on a grid and export to VTK#
Evaluate the saved fields on a grid and export the result to VTK format.
[6]:
# Export the results to VTK using PostProcessManager
from psydac.api.postprocessing import PostProcessManager
Pm = PostProcessManager(
domain=Omega,
space_file=f'results_regularized_curlcurl/space_info_{Omega.name}.yml',
fields_file=f'results_regularized_curlcurl/field_info_{Omega.name}.h5',
comm=comm
)
Pm.export_to_vtk(
f'results_regularized_curlcurl/visu_{Omega.name}',
grid=None,
npts_per_cell=[4,2,4],
fields=save_fields.keys() # evaluate all the fields that were saved
)
Pm.close()
# If this file is run in sequential, then this step will generate a single .vtu file that can be opened with ParaView.
# If it is run in parallel, it generates a .vtu file per process (containing the local grid) and a .pvtu file. To visualize it in ParaView, open the .pvtu file.
Visualization of the solution in ParaView#
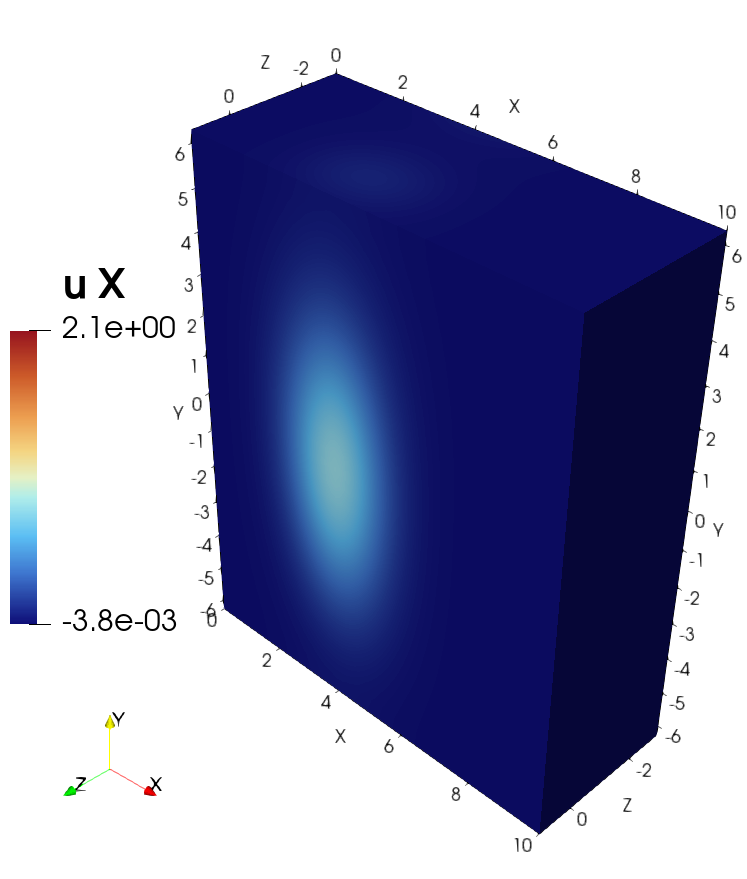
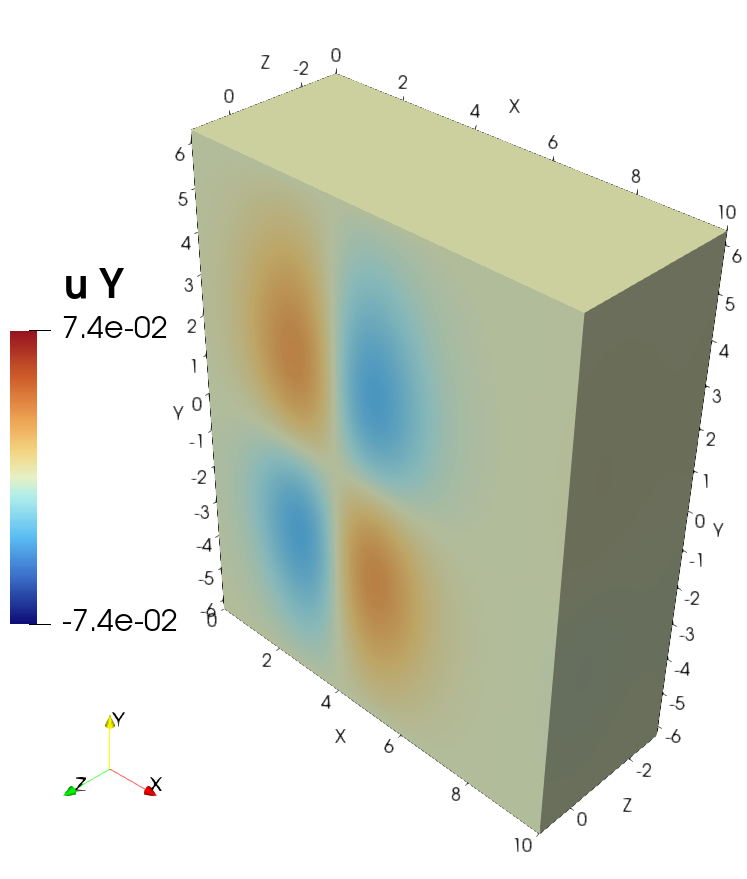
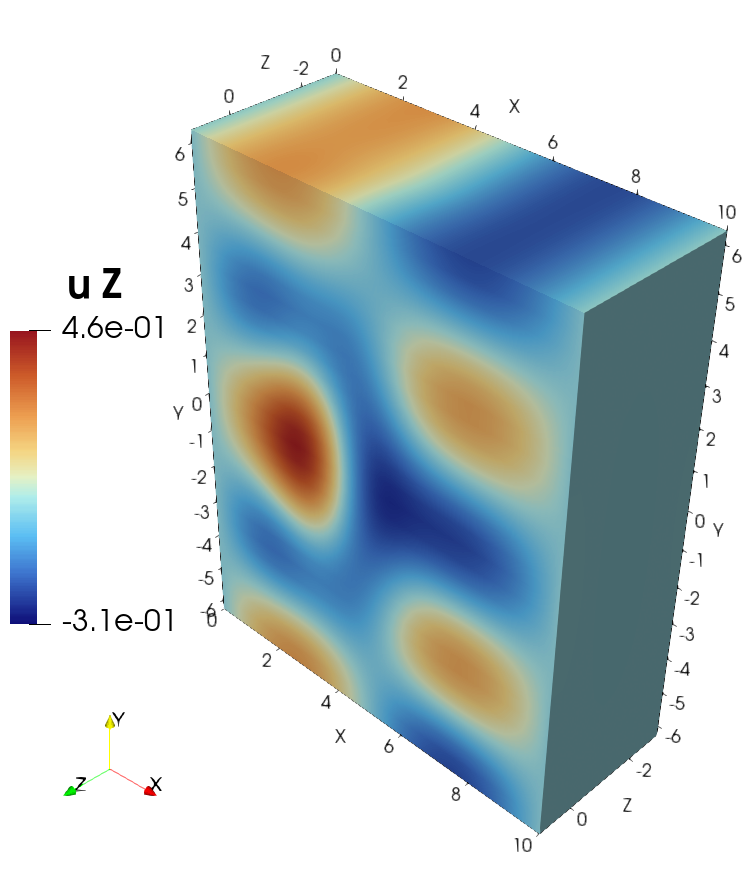
Visualization of the source in ParaView#