Solving Poisson’s equation on non-periodic topological domains#
In this notebook, we will show how to solve poisson’s equation on single patch and multipatch domains. As we are running this in a notebook, everything will be run in serial and hence we are limiting ourselves to a fairly coarse discretization to avoid taking too much time. However, PSYDAC allows for hybrid MPI + OpenMP parallelization with barely any changed to the code. The lines that are impacted by that will be preceeded by their MPI equivalent
Step 1 : Building the domain#
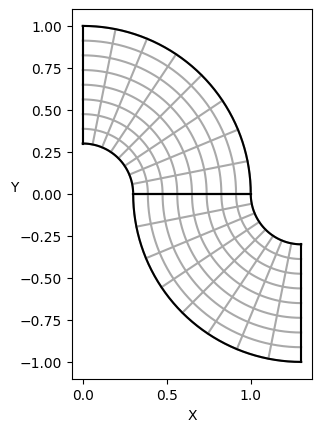
PSYDAC uses the powerful topological tools of SymPDE to build a large variety of domains. Here we show how to do a quarter annulus using a Square and a polar mapping and how to join two annulus to create a multipatch domain. Left commented is an example of a complex multipatch domain built using SymPDE.
[1]:
import numpy as np
from sympde.topology import Square, PolarMapping
from sympde.utilities.utils import plot_domain
from psydac.api.tests.build_domain import build_pretzel
# Define the topological geometry for each patch
rmin, rmax = 0.3, 1.
# First quarter annulus
domain_log_1 = Square('A_1', bounds1=(0., 1.), bounds2=(0., np.pi/2))
F_1 = PolarMapping('F_1', dim=2, c1=0., c2=0., rmin=rmin, rmax=rmax)
Omega_1 = F_1(domain_log_1)
# Second quarter annulus
domain_log_2 = Square('A_2', bounds1=(0., 1.), bounds2=(np.pi, 3/2 * np.pi))
F_2 = PolarMapping('F_2', dim=2, c1=rmin+rmax, c2=0., rmin=rmax, rmax=rmin)
Omega_2 = F_2(domain_log_2)
# Join the patches
from sympde import Domain
connectivity = [((0,1,-1),(1,1,-1), 1)]
patches = [Omega_1, Omega_2]
Omega = Domain.join(patches, connectivity, 'domain')
# Example of a complex multi-patch domain
# Omega = build_pretzel()
# Simple visualization of the topological domain
plot_domain(Omega, draw=False, isolines=True)
Step 2: Defining the Abstract PDE model using SymPDE#
[2]:
from sympde.calculus import grad, dot
from sympde.calculus import minus, plus
from sympde.expr.expr import LinearForm, BilinearForm
from sympde.expr.expr import integral
from sympde.expr.expr import Norm
from sympde.expr import find, EssentialBC
from sympde.topology import ScalarFunctionSpace
from sympde.topology import elements_of
from sympde.topology import NormalVector
# Define the abstract model to solve Poisson's equation using the manufactured solution method
x,y = Omega.coordinates
solution = x**2 + y**2
f = -4
V = ScalarFunctionSpace('V', Omega, kind=None)
u, v = elements_of(V, names='u, v')
nn = NormalVector('nn')
bc = EssentialBC(u, solution, Omega.boundary)
error = u - solution
I = Omega.interfaces
kappa = 10**3
expr_I =- 0.5*dot(grad(plus(u)),nn)*minus(v) + 0.5*dot(grad(minus(v)),nn)*plus(u) - kappa*plus(u)*minus(v)\
+ 0.5*dot(grad(minus(u)),nn)*plus(v) - 0.5*dot(grad(plus(v)),nn)*minus(u) - kappa*plus(v)*minus(u)\
- 0.5*dot(grad(minus(v)),nn)*minus(u) - 0.5*dot(grad(minus(u)),nn)*minus(v) + kappa*minus(u)*minus(v)\
+ 0.5*dot(grad(plus(v)),nn)*plus(u) + 0.5*dot(grad(plus(u)),nn)*plus(v) + kappa*plus(u)*plus(v)
expr = dot(grad(u),grad(v))
a = BilinearForm((u,v), integral(Omega, expr) + integral(I, expr_I))
l = LinearForm(v, integral(Omega, f*v))
equation = find(u, forall=v, lhs=a(u,v), rhs=l(v), bc=bc)
l2norm = Norm(error, Omega, kind='l2')
h1norm = Norm(error, Omega, kind='h1')
Step 3: Discretizing the domain, spaces and equations#
[3]:
# Discretize the geometry and equation
from psydac.api.discretization import discretize
from psydac.api.settings import PSYDAC_BACKENDS
# The backends are as follows and can all be run with OpenMP by setting backend['omp'] = True.
# For all but the Python backend, compilation flags can be accessed and changed using backend['flags'].
# - Python backend: Generate and runs python files. Accessed via PSYDAC_BACKENDS['python']
# - Pyccel GCC backend: Accessed via PSYDAC_BACKENDS['pyccel-gcc']
# - Pyccel Intel backend: Accessed via PSYDAC_BACKENDS['pyccel-intel']
# - Pyccel PGI backend: Accessed via PSYDAC_BACKENDS['pyccel-pgi']
# - Pyccel Numba backend: Accessed via PSYDAC_BACKENDS['numba']
backend = PSYDAC_BACKENDS['python']
# Uncomment to use OpenMp
# import os
# os.environ['OMP_NUM_THREADS'] = "4"
# backend['omp'] = True
ncells = [10, 10]
degree = [2, 2]
periodic = [False, False]
nquads = [p + 1 for p in degree]
# MPI version
# from mpi4py import MPI
# comm = MPI.COMM_WORLD
# Omega_h = discretize(Omega, ncells=ncells, comm=comm)
Omega_h = discretize(Omega, ncells=ncells, periodic=periodic)
Vh = discretize(V, Omega_h, degree=degree)
equation_h = discretize(equation, Omega_h, [Vh, Vh], nquads=nquads, backend=backend)
l2norm_h = discretize(l2norm, Omega_h, Vh, nquads=nquads, backend=backend)
h1norm_h = discretize(h1norm, Omega_h, Vh, nquads=nquads, backend=backend)
Step 4: Solving the equation and computing the error norms#
[4]:
# Set the solver parameters
# 'cg' -> Conjugate gradient method
equation_h.set_solver('cg', info=True, tol=1e-14)
import time
t0_s = time.time()
uh, info = equation_h.solve()
t1_s = time.time()
t0_d = time.time()
l2_error = l2norm_h.assemble(u=uh)
h1_error = h1norm_h.assemble(u=uh)
t1_d = time.time()
print( '> CG info :: ',info )
print( '> L2 error :: {:.2e}'.format(l2_error))
print( '> H1 error :: {:.2e}'.format(h1_error))
print( '> Solution time :: {:.2e}s'.format(t1_s - t0_s))
print( '> Evaluat. time :: {:.2e}s '.format(t1_d - t0_d))
> CG info :: {'niter': 108, 'success': True, 'res_norm': 4.8188464197114785e-15}
> L2 error :: 2.17e-05
> H1 error :: 1.44e-03
> Solution time :: 1.40e+00s
> Evaluat. time :: 5.12e-02s
Step 5: Saving the results#
[5]:
# Save the results using OutputManager
from psydac.api.postprocessing import OutputManager
import os
os.makedirs('results_poisson', exist_ok=True)
Om = OutputManager(
f'results_poisson/space_info_{Omega.name}',
f'results_poisson/field_info_{Omega.name}',
# MPI version
# comm=comm,
)
Om.add_spaces(V=Vh)
Om.export_space_info()
Om.set_static()
Om.export_fields(u=uh)
Om.close()
Step 6: Exporting the results to VTK and visualizing them with Paraview#
[6]:
# Export the results to VTK using PostProcessManager
from psydac.api.postprocessing import PostProcessManager
from sympy import lambdify
Pm = PostProcessManager(
domain=Omega,
space_file=f'results_poisson/space_info_{Omega.name}.yaml',
fields_file=f'results_poisson/field_info_{Omega.name}.h5',
# MPI version
# comm=comm,
)
Pm.export_to_vtk(
f'results_poisson/visu_{Omega.name}',
grid=None,
npts_per_cell=3,
fields='u',
additional_physical_functions={'exact_solution': lambdify(Omega.coordinates, solution, modules='numpy')}
)
Pm.close()
Example: Results on the pretzel domain#
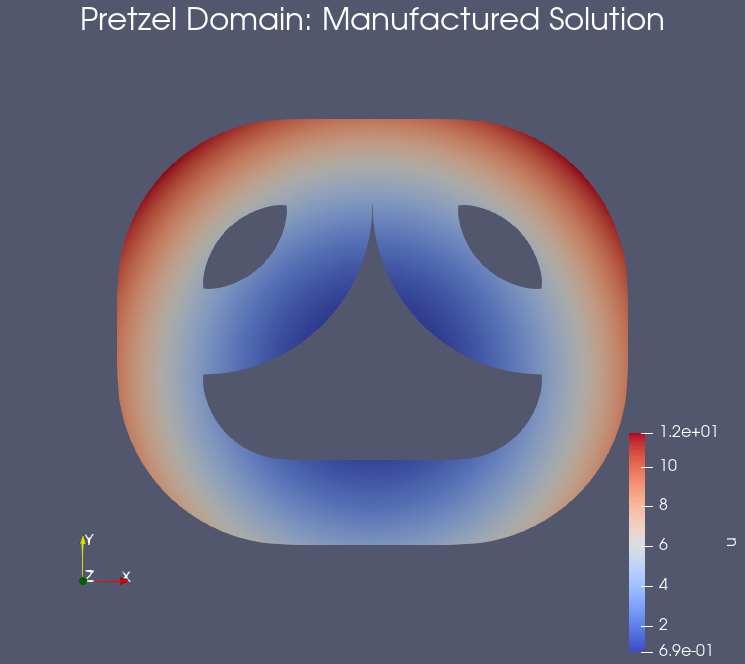
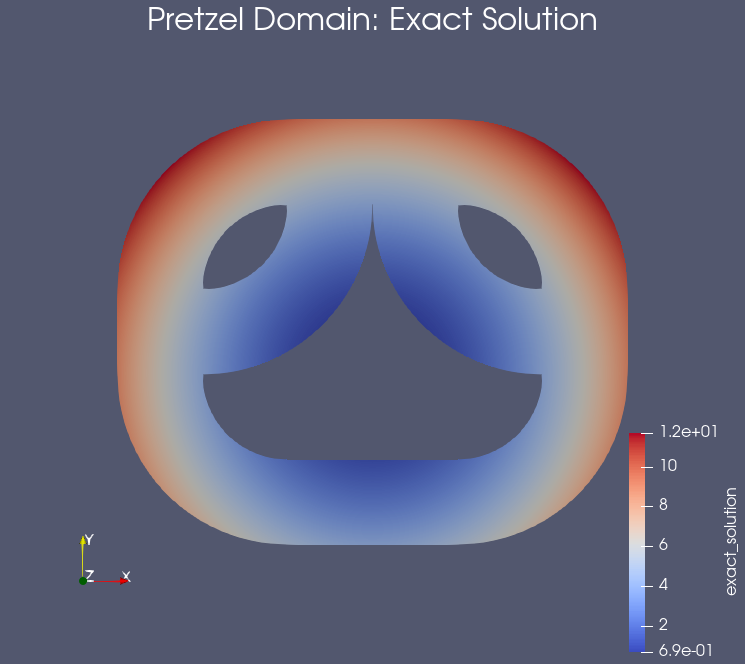
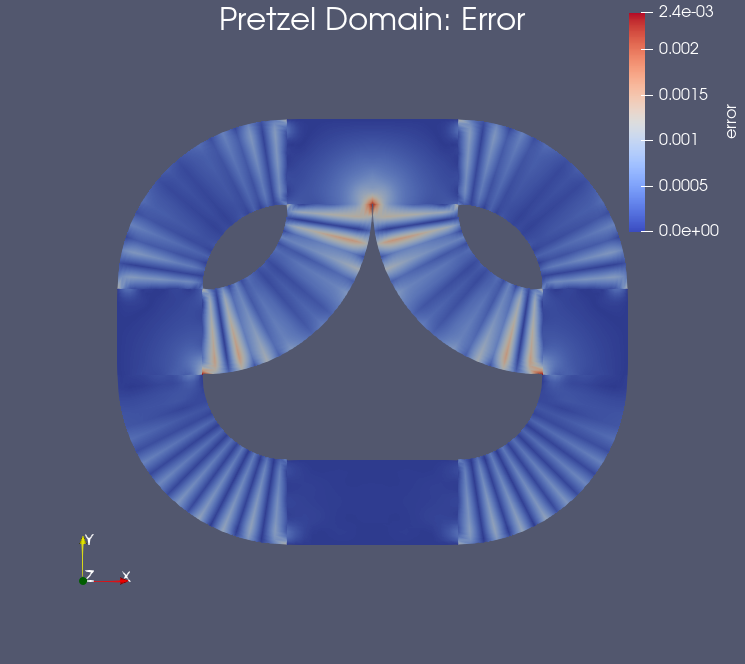
Testing the notebook#
[7]:
import ipytest
ipytest.autoconfig(raise_on_error=True)
[8]:
%%ipytest
def test_l2error():
assert l2_error < 3e-05
def test_h1error():
assert h1_error < 2e-03
.. [100%]
2 passed in 0.02s